Deep Learning-Based Blood Cell Image Classification Using ResNet18 Architecture
DOI:
https://doi.org/10.56705/ijodas.v6i2.300Keywords:
Blood Cell Classification, ResNet18, Deep Learning, Medical Imaging, Multi-class EvaluationAbstract
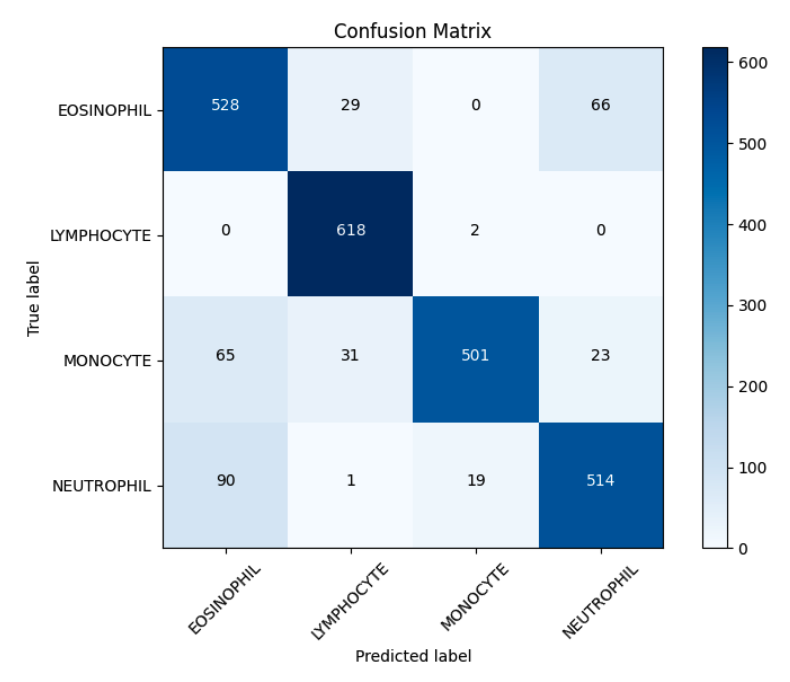
The classification of white blood cells (WBC) plays a critical role in haematological diagnostics, yet manual examination remains a labour-intensive and subjective process. In response to this challenge, this study investigates the application of deep learning, specifically the ResNet18 convolutional neural network architecture, for the automated classification of blood cell images into four classes: eosinophils, lymphocytes, monocytes, and neutrophils. The dataset used comprises microscopic images annotated by cell type and is divided into training and validation sets with an 80:20 ratio. Standard pre-processing techniques such as image normalization and augmentation were applied to enhance model robustness and generalization. The model was fine-tuned using transfer learning with pre-trained weights from ImageNet and optimized using the Adam optimizer. Performance was evaluated through a comprehensive set of metrics including accuracy, precision, recall, F1-score, mean squared error (MSE), and root mean squared error (RMSE). The best model achieved a validation accuracy of 86.89%, with macro-averaged precision, recall, and F1-score of 0.8738, 0.8690, and 0.8688, respectively. Lymphocyte classification yielded the highest F1-score (0.9515), while eosinophils posed the greatest classification challenge, as evidenced by lower precision and higher misclassification rates in the confusion matrix. Error-based evaluation further supported the model’s consistency, with an MSE of 0.7125 and RMSE of 0.8441. These results confirm that ResNet18 is capable of learning discriminative features in complex haematological imagery, providing an efficient and reliable alternative to manual analysis. The findings suggest potential for practical implementation in clinical workflows and pave the way for further research involving multi-model ensembles or cell segmentation pre-processing for improved precision
Downloads
References
H. Liu, M. Zhao, C. She, H. Peng, M. Liu, and B. Li, “Classification of CT scan and X-ray dataset based on deep learning and particle swarm optimization,” PLoS One, vol. 20, no. 1, p. e0317450, Jan. 2025, doi: 10.1371/journal.pone.0317450.
H. Jin, K. Han, H. Xia, B. Xu, and X. Jin, “Detection of weeds in vegetables using image classification neural networks and image processing,” Front. Phys., vol. 13, Jan. 2025, doi: 10.3389/fphy.2025.1496778.
C. Ramathal et al., “Deep learning-driven morphology analysis enables label-free classification of therapeutic agentnaive versus resistant cancer cells.” Jan. 25, 2025, doi: 10.1101/2025.01.22.634357.
Z. Gao, Y. Tian, S.-C. Lin, and J. Lin, “A CT Image Classification Network Framework for Lung Tumors Based on Pre-trained MobileNetV2 Model and Transfer learning, And Its Application and Market Analysis in the Medical Field,” Appl. Comput. Eng., vol. 133, no. 1, pp. 90–96, Jan. 2025, doi: 10.54254/2755-2721/2025.20605.
A. K. Mali, S. Murugappan, J. R. Prasad, S. A. M. Tofail, and N. D. Thorat, “A Deep Learning Pipeline for Morphological and Viability Assessment of 3D Cancer Cell Spheroids.” Jan. 24, 2025, doi: 10.1101/2025.01.20.633939.
L. Ma, “Multi-Plant Disease Identification Based on Lightweight ResNet18 Model,” Agronomy, vol. 13, no. 11, 2023, doi: 10.3390/agronomy13112702.
S. She, “Evaluation of Defects Depth for Metal Sheets Using Four-Coil Excitation Array Eddy Current Sensor and Improved ResNet18 Network,” IEEE Sens. J., vol. 24, no. 12, pp. 18955–18967, 2024, doi: 10.1109/JSEN.2024.3367816.
S. T. Prasath, “Deep Learning Framework for Colorectal Cancer Classification Using ResNet18 Based on Dietary Habits Related to Meat Intake and Cooking Methods,” IEEE Access, vol. 12, pp. 99453–99468, 2024, doi: 10.1109/ACCESS.2024.3430036.
P. Maheswari, “MangoYieldNet: Fruit yield estimation for mango orchards using DeepLabv3 + with ResNet18 architecture,” Multimed. Tools Appl., 2025, doi: 10.1007/s11042-025-20791-5.
J. Wang, “Pipeline Landmark Classification of Miniature Pipeline Robot π-II Based on Residual Network ResNet18,” Machines, vol. 12, no. 8, 2024, doi: 10.3390/machines12080563.
S. Liu, “Fault Diagnosis Strategy Based on BOA-ResNet18 Method for Motor Bearing Signals with Simulated Hydrogen Refueling Station Operating Noise,” Appl. Sci., vol. 14, no. 1, 2024, doi: 10.3390/app14010157.
A. Gupta and S. Gupta, “Enhanced Classification of Imbalanced Medical Datasets using Hybrid Data-Level, Cost-Sensitive and Ensemble Methods,” Int. Res. J. Multidiscip. Technovation, pp. 58–76, Apr. 2024, doi: 10.54392/irjmt2435.
A. Sinra, B. S. W. Poetro, H. Angriani, H. Zein, and ..., “Optimizing Neurodegenerative Disease Classification with Canny Segmentation and Voting Classifier: An Imbalanced Dataset Study,”.
Frieyadie, M. Abdullah, and F. A. Setiawan, “The Effectiveness of Resampling Method for Handling Class Imbalances in Software Defect Prediction,” in 2023 International Conference on Information Technology Research and Innovation (ICITRI), Aug. 2023, pp. 22–27, doi: 10.1109/ICITRI59340.2023.10249255.
J. Sharma, “Deep Learning Approach for Poultry Disease Classification and Early Detection: ResNet18,” 2025 International Conference on Pervasive Computational Technologies, ICPCT 2025. pp. 139–143, 2025, doi: 10.1109/ICPCT64145.2025.10940397.
W. Yang, “Optimizing Facial Expression Recognition: A One-Class Classification Approach Using ResNet18 and CBAM,” Proceedings - 2024 3rd International Conference on Computer Technologies, ICCTech 2024. pp. 1–5, 2024, doi: 10.1109/ICCTech61708.2024.00009.
J. Zhang, “Prediction of Intrinsically Disordered Proteins Based on Deep Neural Network-ResNet18,” C. - Comput. Model. Eng. Sci., vol. 131, no. 2, pp. 905–917, 2022, doi: 10.32604/cmes.2022.019097.
W. Zhang, “Identifying Multiple Apple Leaf Diseases Based on the Improved CBAM-ResNet18 Model under Weak Supervision,” Smart Agric., vol. 5, no. 1, pp. 111–121, 2023, doi: 10.12133/j.smartag.SA202301005.
Y. Jia, “Research on Improving ResNet18 for Classifying Complex Images Based on Attention Mechanism,” Communications in Computer and Information Science, vol. 2139. pp. 123–139, 2024, doi: 10.1007/978-981-97-3948-6_13.
S. Cheng, “Efficient acne classification based on Resnet18,” Proceedings of SPIE - The International Society for Optical Engineering, vol. 13545. 2025, doi: 10.1117/12.3060304.
I. Tahyudin, “High-Accuracy Stroke Detection System Using a CBAM-ResNet18 Deep Learning Model on Brain CT Images,” J. Appl. Data Sci., vol. 6, no. 1, pp. 788–799, 2025, doi: 10.47738/jads.v6i1.569.
H. S. Obaid, S. A. Dheyab, and S. S. Sabry, “The Impact of Data Pre-Processing Techniques and Dimensionality Reduction on the Accuracy of Machine Learning,” in 2019 9th Annual Information Technology, Electromechanical Engineering and Microelectronics Conference (IEMECON), Mar. 2019, pp. 279–283, doi: 10.1109/IEMECONX.2019.8877011.
G. Pallavi, “Brain tumor detection with high accuracy using random forest and comparing with thresholding method,” AIP Conference Proceedings, vol. 2853, no. 1. 2024, doi: 10.1063/5.0198189.
T. Hastie, R. Tibshirani, and J. Friedman, The Elements of Statistical Learning Data Mining, Inference, and Prediction. 2009.
R. S. Ram, M. V. Kumar, T. M. Al-Shami, M. Masud, H. Aljuaid, and M. Abouhawwash, “Deep Fake Detection Using Computer Vision-Based Deep Neural Network with Pairwise Learning,” Intell. Autom. Soft Comput., vol. 35, no. 2, pp. 2449–2462, 2023, doi: 10.32604/iasc.2023.030486.
Downloads
Published
Issue
Section
License
Authors retain copyright and full publishing rights to their articles. Upon acceptance, authors grant Indonesian Journal of Data and Science a non-exclusive license to publish the work and to identify itself as the original publisher.
Self-archiving. Authors may deposit the submitted version, accepted manuscript, and version of record in institutional or subject repositories, with citation to the published article and a link to the version of record on the journal website.
Commercial permissions. Uses intended for commercial advantage or monetary compensation are not permitted under CC BY-NC 4.0. For permissions, contact the editorial office at ijodas.journal@gmail.com.
Legacy notice. Some earlier PDFs may display “Copyright © [Journal Name]” or only a CC BY-NC logo without the full license text. To ensure clarity, the authors maintain copyright, and all articles are distributed under CC BY-NC 4.0. Where any discrepancy exists, this policy and the article landing-page license statement prevail.














