Performance Comparison of MobileNetV2 and NASNetMobile Architectures in Soybean Leaf Disease Classification
DOI:
https://doi.org/10.56705/ijodas.v6i2.243Keywords:
Deep Learning, Image Classification, MobileNetV2, NASNetMobile, Soybean Leaf DiseaseAbstract
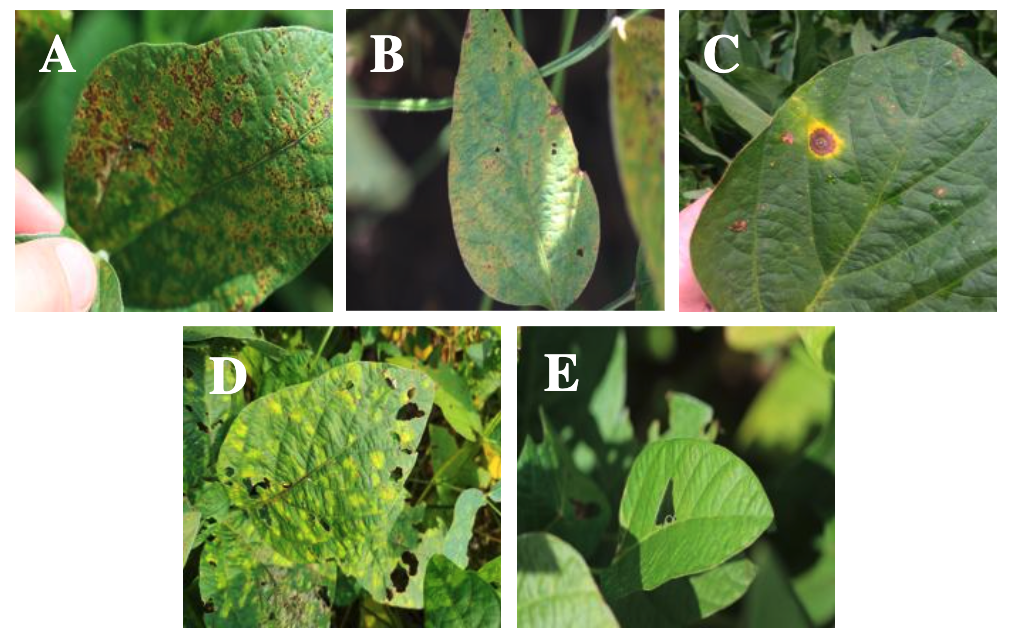
Soybean is one of the essential commodities in Indonesia, commonly used as a raw material for tofu and tempeh, making it highly sought after. However, soybean production has decreased by up to 30% due to disease attacks, necessitating preventive measures. This study aims to compare two Convolutional Neural Network (CNN) architectures, MobileNetV2 and NASNetMobile, in classifying soybean leaf diseases. The models were trained using a leaf image dataset collected directly from agricultural fields and categorized into five classes. The dataset underwent augmentation to increase its size, resulting in a total of 6,000 images, which were then split with an 80:10:10 ratio. The models were trained using the Adam optimizer with a learning rate of 0.001, optimized using ReduceLROnPlateau, and a dropout rate of 0.2 to prevent overfitting. Evaluation results using a confusion matrix indicated that MobileNetV2 performed better with an accuracy of 96.67%, precision of 96.70%, recall of 96.67%, and an F1-score of 96.68%, compared to NASNetMobile, which achieved an accuracy of 86.33%, precision of 86.91%, recall of 86.33%, and an F1-score of 86.40%.
Downloads
References
R. Krisdiana, N. Prasetiaswati, I. Sutrisno, F. Rozi, A. Harsono, and M. J. Mejaya, “Financial feasibility and competitiveness levels of soybean varieties in rice-based cropping system of Indonesia,” Sustain., vol. 13, no. 15, 2021, doi: 10.3390/su13158334.
D. R. Sari, “Analysis of Factors Affecting Chili Imports in Indonesia in 2000-2014,” vol. 4, no. 4, pp. 1555–1565, 2016.
A. P. Ratri, Y. Susanti, and I. Slamet, “The Factors Affecting Soybean Production in Indonesia Using Robust Regression with Least Median of Squares (LMS) Estimation,” vol. 2021, no. 2020, pp. 70–78, 2021, doi: 10.11594/nstp.2021.1110.
G. W. A. Susanto et al., “Stability Analysis to Select the Stable and High Yielding of Black Soybean (Glycine max (L) Merril) in Indonesia,” Int. J. Agron., vol. 2023, 2023, doi: 10.1155/2023/7255444.
M. Borah and B. Deb, “A Review on Symptomatology, Epidemiology and Integrated Management Strategies of Some Economically Important Fungal Diseases of Soybean (Glycine max),” Int. J. Curr. Microbiol. Appl. Sci., vol. 9, no. 11, pp. 1247–1267, 2020, doi: 10.20546/ijcmas.2020.911.147.
J. Kim, S. H. Ahn, J. S. Yang, S. Choi, H. W. Jung, and J. Jeon, “Plant Protective and Growth Promoting Effects of Seed Endophytes in Soybean Plants,” Plant Pathol. J., vol. 39, no. 5, pp. 513–521, 2023, doi: 10.5423/PPJ.OA.06.2023.0091.
J. Patel et al., “Deciphering genetic factors contributing to enhanced resistance against Cercospora leaf blight in soybean (Glycine max L.) using GWAS analysis,” Front. Genet., vol. 15, no. May, pp. 1–13, 2024, doi: 10.3389/fgene.2024.1377223.
M. M. Khalid and O. Karan, “Deep Learning for Plant Disease Detection,” Int. J. Math. Stat. Comput. Sci., vol. 2, pp. 75–84, 2023, doi: 10.59543/ijmscs.v2i.8343.
R. Rashid, W. Aslam, R. Aziz, and G. Aldehim, “An Early and Smart Detection of Corn Plant Leaf Diseases Using IoT and Deep Learning Multi-Models,” IEEE Access, vol. 12, pp. 23149–23162, 2024, doi: 10.1109/ACCESS.2024.3357099.
M. K. Nichat and S. E. Yedey, “Deep Learning Techniques for Identification of Different Malvaceae Plant Leaf Diseases,” EAI Endorsed Trans. Internet Things, vol. 10, pp. 1–10, 2024, doi: 10.4108/eetiot.5394.
O. Iparraguirre-Villanueva et al., “Disease Identification in Crop Plants based on Convolutional Neural Networks,” Int. J. Adv. Comput. Sci. Appl., vol. 14, no. 3, pp. 519–528, 2023, doi: 10.14569/IJACSA.2023.0140360.
S. Ghazal, A. Munir, and W. S. Qureshi, “Computer vision in smart agriculture and precision farming: Techniques and applications,” Artif. Intell. Agric., vol. 13, pp. 64–83, 2024, doi: 10.1016/j.aiia.2024.06.004.
K. Mahadevan, A. Punitha, and J. Suresh, “A Real Time Performance Comparison of Rice Plant Disease Identification System using Deep CNN Models,” Int. Res. J. Multidiscip. Scope, vol. 5, no. 3, pp. 249–258, 2024, doi: 10.47857/irjms.2024.v05i03.0159.
R. Li et al., “Enhancing Wheat Spike Counting and Disease Detection Using a Probability Density Attention Mechanism in Deep Learning Models for Precision Agriculture,” Plants, vol. 13, no. 24, 2024, doi: 10.3390/plants13243462.
S. E. Arnaud, N. Rehema, S. Aoki, and M. L. Kananu, “Comparison of Deep Learning Architectures for Late Blight and Early Blight Disease Detection on Potatoes,” Open J. Appl. Sci., vol. 12, no. 05, pp. 723–743, 2022, doi: 10.4236/ojapps.2022.125049.
P. K. Das, S. S. Rupa, S. Pumrin, U. C. Das, and M. K. Hossen, “Deep Learning for Plant Disease Detection and Classification: A Systematic Analysis and Review,” Curr. Appl. Sci. Technol., vol. 24, no. 4, 2024, doi: 10.55003/cast.2024.259016.
Y. Ferdi, “Data Augmentation through Background Removal for Apple Leaf Disease Classification Using the MobileNetV2 Model”.
M. Rafid, A. Luthfiarta, M. Naufal, M. D. Al Fahreza, and M. Indrawan, “The Effect of LAB Color Space with NASNetMobile Fine-tuning on Model Performance for Crowd Detection,” Adv. Sustain. Sci. Eng. Technol., vol. 6, no. 1, pp. 1–9, 2024, doi: 10.26877/asset.v6i1.17821.
L. Barany, N. Hore, A. Stadlbauer, M. Buchfelder, and S. Brandner, “Prediction of the Topography of the Corticospinal Tract on T1-Weighted MR Images Using Deep-Learning-Based Segmentation,” Diagnostics, vol. 13, no. 5, pp. 1–13, 2023, doi: 10.3390/diagnostics13050911.
S. Ray, K. Alshouiliy, and D. P. Agrawal, “Dimensionality reduction for human activity recognition using google colab,” Inf., vol. 12, no. 1, pp. 1–23, 2021, doi: 10.3390/info12010006.
R. Khadka, P. G. Lind, A. Yazidi, and A. Belhadi, “DREAMS: A python framework to train deep learning models with model card reporting for medical and health applications,” pp. 1–13, 2024.
N. A. Lafta and Z. A. A. Abbood, “Comprehensive Review and Comparative Analysis of Keras for Deep Learning Applications: A Survey on Face Detection Using Convolutional Neural Networks,” Int. J. Relig., vol. 5, no. 11, pp. 1203–1213, 2024, doi: 10.61707/gkh1m822.
A. Al-Adwan, “Evaluating the Effectiveness of Brain Tumor Image Generation using Generative Adversarial Network with Adam Optimizer,” Int. J. Adv. Comput. Sci. Appl., vol. 15, no. 6, pp. 512–521, 2024, doi: 10.14569/IJACSA.2024.0150653.
A. L. C. Ottoni, R. M. de Amorim, M. S. Novo, and D. B. Costa, “Tuning of data augmentation hyperparameters in deep learning to building construction image classification with small datasets,” Int. J. Mach. Learn. Cybern., vol. 14, no. 1, pp. 171–186, 2023, doi: 10.1007/s13042-022-01555-1.
I. P. Kamila, C. A. Sari, E. H. Rachmawanto, and N. R. D. Cahyo, “A Good Evaluation Based on Confusion Matrix for Lung Diseases Classification using Convolutional Neural Networks,” Adv. Sustain. Sci. Eng. Technol., vol. 6, no. 1, pp. 1–8, 2024, doi: 10.26877/asset.v6i1.17330.
Downloads
Published
Issue
Section
License
Authors retain copyright and full publishing rights to their articles. Upon acceptance, authors grant Indonesian Journal of Data and Science a non-exclusive license to publish the work and to identify itself as the original publisher.
Self-archiving. Authors may deposit the submitted version, accepted manuscript, and version of record in institutional or subject repositories, with citation to the published article and a link to the version of record on the journal website.
Commercial permissions. Uses intended for commercial advantage or monetary compensation are not permitted under CC BY-NC 4.0. For permissions, contact the editorial office at ijodas.journal@gmail.com.
Legacy notice. Some earlier PDFs may display “Copyright © [Journal Name]” or only a CC BY-NC logo without the full license text. To ensure clarity, the authors maintain copyright, and all articles are distributed under CC BY-NC 4.0. Where any discrepancy exists, this policy and the article landing-page license statement prevail.














